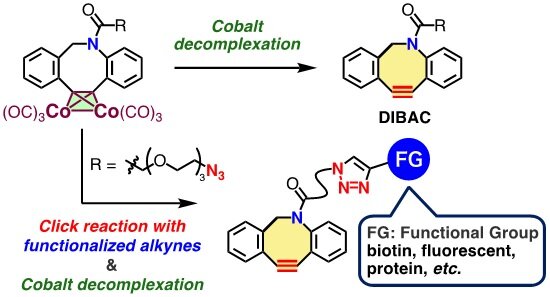
Facile synthesis of functionalized dibenzo-fused azacyclooctynes (DIBACs) by click reaction and subsequent cobalt decomplexation. Credit: Laboratory of Chemical Bioscience, TMDU
The 2022 Nobel Prize in Chemistry was awarded in part for what can be a quite difficult problem: precisely altering one aspect of biomolecules without affecting the rest of the cell. Now, in a study recently published in Organic Letters, researchers from Tokyo Medical and Dental University (TMDU) and coworkers have concisely synthesized a class of molecules that will greatly facilitate such work.
Imagine experimenting with the preparation of bread by neglecting to add yeast. You’ll notice many effects—in addition to a failure to rise, the bread’s flavor and aroma will change. In other words, by changing one aspect of the recipe, you’ve fundamentally changed the entire end result. You can envision living cells as being like a vastly more complex bread, with many ingredients that work together. To study any specific aspect of cellular function you need precise tools that don’t affect any of the other chemistry that’s going on in the cell.
Strain-promoted azide–alkyne cycloaddition (SPAAC) reactions that use dibenzoazacyclooctynes (DIBACs) enable precise biomolecular analysis. Expanding the chemical diversity of DIBACs would therefore correspondingly expand the possibilities for SPAAC reactions and would help unlock the vast potential that this chemical reaction presents. Thus, the researchers chose to focus on this area in an effort to develop a new, efficient, and streamlined method for creating synthetic DIBACs that could be used in SPAAC reactions.
“It is generally difficult to prepare anything but simple DIBAC molecules,” explains Yuki Sakata, lead author. “Accordingly, we focused on synthesizing diverse DIBACs by a versatile procedure.”
A critical step of the present work was concise synthesis of a cobalt complex that enabled straightforward synthesis of the desired cycloalkyne—the chemical unit that’s essential for DIBAC’s function in biomolecular analysis by SPAAC. For example, one DIBAC was obtained in six steps with a 72% yield; the previous method required eight steps and gave only a 45% yield. Furthermore, even highly complex DIBACs that were incompatible with previous synthetic approaches were obtained through this new method.
“We used DIBAC-enabled SPAAC chemistry to modify an azido-HaloTag protein, which is a versatile tool in protein function and interaction studies,” says Takamitsu Hosoya, senior author. “Applications in otherwise complex biomolecular functionalizations are underway in our laboratory.”
This work succeeded in expanding the range of chemistry that’s available to practitioners of SPAAC chemistry. The researchers’ facile DIBAC synthesis protocol will be useful for ongoing efforts at interrogating cellular function in ways that are precise, gentle, and nondisruptive to overall cellular physiology.
More information: Yuki Sakata et al, Synthesis of Functionalized Dibenzoazacyclooctynes by a Decomplexation Method for Dibenzo-Fused Cyclooctyne–Cobalt Complexes, Organic Letters (2022). DOI: 10.1021/acs.orglett.2c03832
Provided by Tokyo Medical and Dental University
Citation: Biomolecular analyses now have an expanded chemical toolkit (2023, January 17) retrieved 29 January 2023 from https://phys.org/news/2023-01-biomolecular-analyses-chemical-toolkit.html
This document is subject to copyright. Apart from any fair dealing for the purpose of private study or research, no part may be reproduced without the written permission. The content is provided for information purposes only.